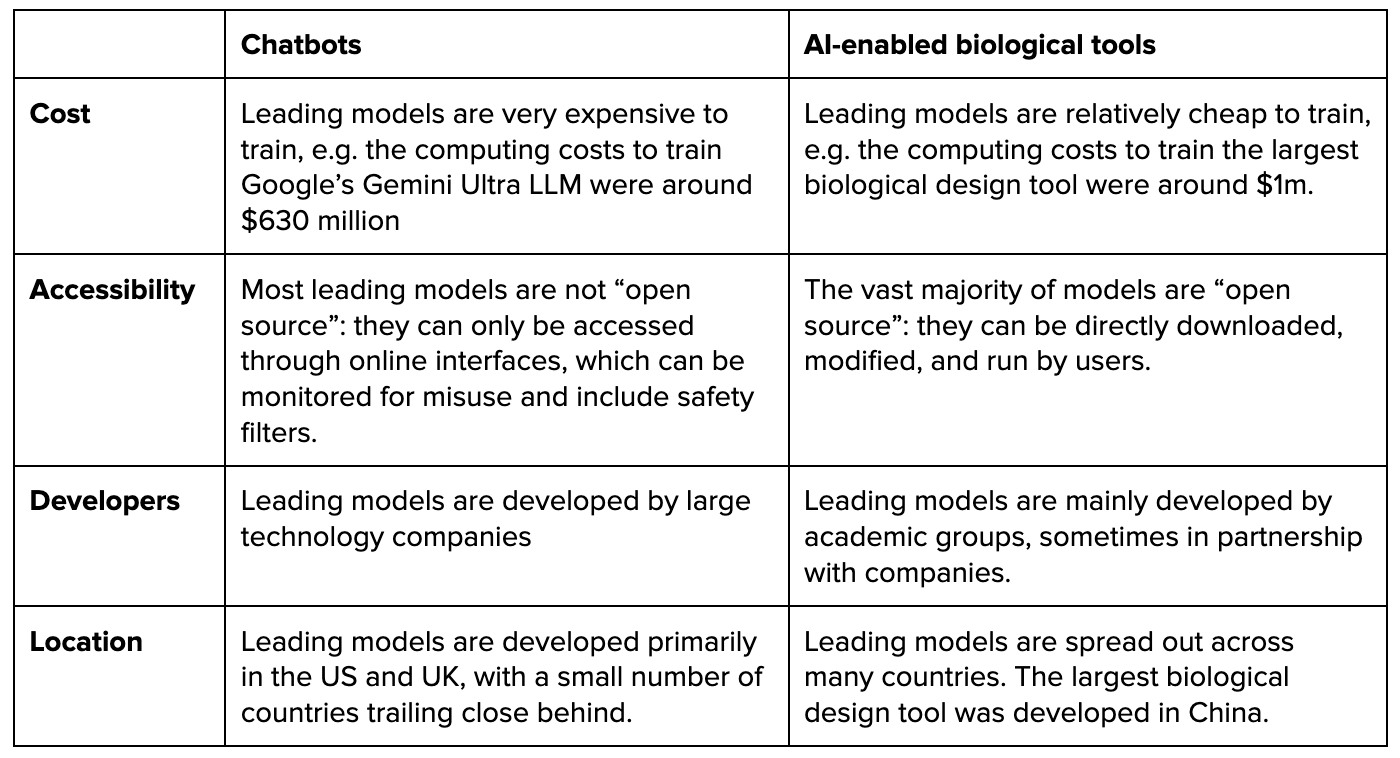
Previously two options were available JSON Mode & Function Calling…
Enabling OpenAI’s JSON mode doesn’t ensure that the output will adhere to a specific predefined JSON schema. It only guarantees that the JSON will be valid and parse without errors.
The challenge with OpenAI’s JSON Mode lies in the significant variability of the JSON output with each inference, making it impossible to predefine a consistent JSON schema.
To clarify, the chat completion API itself doesn’t call any functions, but the model can generate JSON output that you can use in your code to trigger function calls.
Last year OpenAI introduced JSON mode as a valuable tool for developers aiming to build reliable applications using their models.
Although JSON mode enhances the model’s ability to generate valid JSON outputs, but has I have highlighted in previous articles, it does not ensure that the responses will adhere to a specific schema. Which makes this a more experimental feature than a production ready feature.
Now, OpenAI is introducing Structured Outputs in the API, a new feature designed to guarantee that model-generated outputs will precisely match the JSON Schemas provided by developers.
Structured output is available in two formats, Function Calling & A new option for the response_format parameter.
The following Python code of a Function Calling example can be copied and pasted as-is into a notebook:
# Install the requests library if not already installed
!pip install requestsimport requests
import json
# Define your OpenAI API key
api_key = ''
# Define the API endpoint
url = "https://api.openai.com/v1/chat/completions"
# Define the headers with the API key
headers = {
"Authorization": f"Bearer {api_key}",
"Content-Type": "application/json"
}
# Define the data for the API request
data = {
"model": "gpt-4o-2024-08-06",
"messages": [
{
"role": "system",
"content": "You are a helpful assistant. The current date is August 6, 2024. You help users query for the data they are looking for by calling the query function."
},
{
"role": "user",
"content": "look up all my orders in may of last year that were fulfilled but not delivered on time"
}
],
"tools": [
{
"type": "function",
"function": {
"name": "query",
"description": "Execute a query.",
"strict": True,
"parameters": {
"type": "object",
"properties": {
"table_name": {
"type": "string",
"enum": ["orders"]
},
"columns": {
"type": "array",
"items": {
"type": "string",
"enum": [
"id",
"status",
"expected_delivery_date",
"delivered_at",
"shipped_at",
"ordered_at",
"canceled_at"
]
}
},
"conditions": {
"type": "array",
"items": {
"type": "object",
"properties": {
"column": {
"type": "string"
},
"operator": {
"type": "string",
"enum": ["=", ">", "=", " },
"value": {
"anyOf": [
{
"type": "string"
},
{
"type": "number"
},
{
"type": "object",
"properties": {
"column_name": {
"type": "string"
}
},
"required": ["column_name"],
"additionalProperties": False
}
]
}
},
"required": ["column", "operator", "value"],
"additionalProperties": False
}
},
"order_by": {
"type": "string",
"enum": ["asc", "desc"]
}
},
"required": ["table_name", "columns", "conditions", "order_by"],
"additionalProperties": False
}
}
}
]
}
# Make the API request
response = requests.post(url, headers=headers, data=json.dumps(data))
# Print the response
print(response.status_code)
print(response.json())
JSON serves as a vital tool for structuring and exchanging data between AI agents and the functions they interact with, ensuring clear, consistent, and reliable communication across various systems and platforms.
✨✨ Follow me on LinkedIn for updates on Large Language Models
I’m currently the Chief Evangelist @ Kore AI. I explore & write about all things at the intersection of AI & language; ranging from LLMs, Chatbots, Voicebots, Development Frameworks, Data-Centric latent spaces & more.
https://openai.com/index/introducing-structured-outputs-in-the-api